Original photo by Polina Tankilevitch from Pexels
What could go wrong if I use float32 instead of float64 ?
I need more speed for my calculation without changing my code alot, is changing data types coud help me?
I have limited RAM for processing huge dataset, what does this post means to me ?
The goal of this post is to answer these question, focusing on speed and precision, without much tough about how it implemented. I’m assuming you to have some familiarity with Python, Numpy and Pandas.
If number is a stick, and variable is a hole. Using a big hole to store a small stick is wasteful. What worse is putting a huge stick to a small hole. Vinson Ciawandy
Numerical Stability
We need to make sure our number calculated correctly. Floating number (in almost every programming language that i know) have some unavoidable inaccuracies. This is the most famous example of such case.
 Visit this website to understand this issue https://0.30000000000000004.com/
Visit this website to understand this issue https://0.30000000000000004.com/
Most of the time, this kind of innacuracies are tolarable. But, there are some cases where this kind of behaviour is unacceptable. Here’s the example of such case :
 Calculating average from a huge pandas series can yield very different result if you use the wrong datatype. That’s why it is important to always do sanity checking to your data and code.
Calculating average from a huge pandas series can yield very different result if you use the wrong datatype. That’s why it is important to always do sanity checking to your data and code.
For float16, the average is nan. Why? After some exploration, i notice that pandas series using np.mean for calculating average and np.mean start with summing all the values and then dividing the sum by count of the values. Since float16 can store number only up to 65504, storing number bigger than that will yield to Inf
import numpy as np
np.array(100000.0,dtype=np.float16)
> array(inf, dtype=float16) # Output
I’m still not sure why calculating mean resulted in nan instead of Inf, but my point still not change that using the wrong data type could led you to wrong result.
Speed
I will conduct several experiment to see the effect of changing datatype. Could it increase the speed? In theory, more precise data type lead to slower calculation time.
- Pandas aggregation
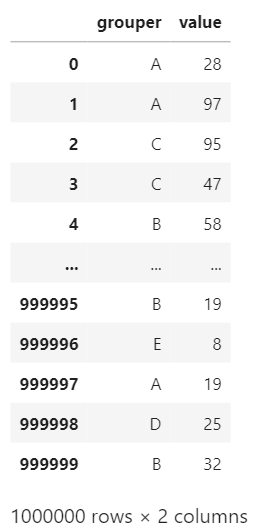
I will create a simple dataframe with one categorical variable and one random integer. I will do several aggregation with different type of data type and see how fast/slow it’s gonna be.
import pandas as pd
import numpy as np
categories = ["A","B","C","D","E"]
length = 1000000
df = pd.DataFrame({"grouper":np.random.choice(categories,length),"value":np.random.randint(0,100,length)})
df
df["value"] = df["value"].astype('float64')
%timeit df.groupby("grouper").agg({"value":["mean","sum","median","std","min","max"]})
df["value"] = df["value"].astype('float32')
%timeit df.groupby("grouper").agg({"value":["mean","sum","median","std","min","max"]})
df["value"] = df["value"].astype('float16')
%timeit df.groupby("grouper").agg({"value":["mean","sum","median","std","min","max"]})
df["value"] = df["value"].astype('int64')
%timeit df.groupby("grouper").agg({"value":["mean","sum","median","std","min","max"]})
df["value"] = df["value"].astype('int32')
%timeit df.groupby("grouper").agg({"value":["mean","sum","median","std","min","max"]})
df["value"] = df["value"].astype('int16')
%timeit df.groupby("grouper").agg({"value":["mean","sum","median","std","min","max"]})
| Data Type | Average Time (ms) | Standard Deviation (ms) | |
|---|---|---|---|
| Float64 | 118 | 2.88 | |
| Float32 | 143 | 2.7 | |
| Float16 | 144 | 6.8 | |
| Int64 | 143 | 4.03 | |
| Int32 | 172 | 6.47 | |
| Int16 | 154 | 2.23 |
Float64 wins the pandas aggregation competition. All experiment run 7 times with 10 loop of repetition.
- Numpy Matrix multiplication
import numpy as np
mat = np.random.randint(0,80,(1000,1000))
mat = mat.astype(np.float64)
%timeit mat.dot(mat)
mat = mat.astype(np.float32)
%timeit mat.dot(mat)
mat = mat.astype(np.float16)
%timeit mat.dot(mat)
mat = mat.astype(np.int64)
%timeit mat.dot(mat)
mat = mat.astype(np.int32)
%timeit mat.dot(mat)
mat = mat.astype(np.int16)
%timeit mat.dot(mat)
| Data Type | Average Time (ms) | Standard Deviation (ms) |
|---|---|---|
| Float64 | 50.1 | 6.28 |
| Float32 | 23 | 2.41 |
| Float16 | 5620 | 611 |
| Int64 | 1560 | 165 |
| Int32 | 1370 | 144 |
| Int16 | 1640 | 210 |
Float32 turn out to be fastest.
Float is faster than integer for this test, but Float16 is really slow compared to the rest. Why? It turns out that float 16 are not supported natively by most processors (What they actually did are emulating float16-like behavior).
Memory
In Python, float data type use more memory than int which is make sense.
from sys import getsizeof
a_float = [float(x) for x in list(range(1,100000000))]
getsizeof(a_float)/1024/1024
>> 819.8971405029297
a_int = list(range(1,100000000))
getsizeof(a_int)/1024/1024
>> 762.9394989013672
To demonstrate the data type effect on pandas, I will use dataset from kaggle competition.
train_df = pd.read_csv('../input/riiid-test-answer-prediction/train.csv', nrows=10**8)
train_df

train_df.info()
>> <class 'pandas.core.frame.DataFrame'>
RangeIndex: 100000000 entries, 0 to 99999999
Data columns (total 10 columns):
# Column Dtype
--- ------ -----
0 row_id int64
1 timestamp int64
2 user_id int64
3 content_id int64
4 content_type_id int64
5 task_container_id int64
6 user_answer int64
7 answered_correctly int64
8 prior_question_elapsed_time float64
9 prior_question_had_explanation object
dtypes: float64(1), int64(8), object(1)
memory usage: 7.5+ GB
The dataset itself use more than 7.5 GB of memory. All the integer stored as int64 and the boolean column store as string/object. Let we fix this by specifying the correct/efficient datatype.
train_df = pd.read_csv('../input/riiid-test-answer-prediction/train.csv',
nrows=10**8,
dtype={'row_id': 'int64',
'timestamp': 'int64',
'user_id': 'int32',
'content_id': 'int16',
'content_type_id': 'int8',
'task_container_id': 'int16',
'user_answer': 'int8',
'answered_correctly': 'int8',
'prior_question_elapsed_time': 'float32',
'prior_question_had_explanation': 'boolean',
})
train_df.info()
>> <class 'pandas.core.frame.DataFrame'>
RangeIndex: 100000000 entries, 0 to 99999999
Data columns (total 10 columns):
# Column Dtype
--- ------ -----
0 row_id int64
1 timestamp int64
2 user_id int32
3 content_id int16
4 content_type_id int8
5 task_container_id int16
6 user_answer int8
7 answered_correctly int8
8 prior_question_elapsed_time float32
9 prior_question_had_explanation boolean
dtypes: boolean(1), float32(1), int16(2), int32(1), int64(2), int8(3)
memory usage: 3.1 GB
Now the dataset become 3.1 GB
Conclusion
When working on small data, the data type is irrelevant since Pandas and Numpy default settings are the best. But when dealing with a huge dataset, we need to give more consideration to which data type we should use because it could save us from mis-precision, long execution time, and huge memory usage.